PhD Thesis - Biochemical, structural and biological analysis of pseudoknot-dependent viral recoding signals
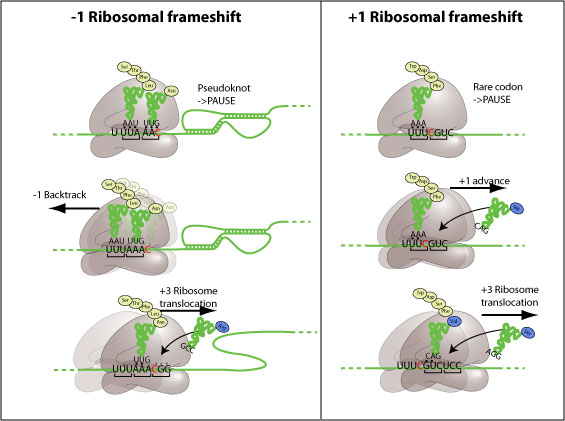 Image credit: ViralZone
Image credit: ViralZone
Summary
Programmed –1 ribosomal frameshifting (-1FS) and stop codon readthrough are used by many RNA viruses and retroviruses to permit the regulated expression of multiple proteins from a single transcript. This dissertation investigated the biological role of readthrough in the model gammaretrovirus murine leukaemia virus (MuLV) and investigated frameshifting at the coronavirus infectious bronchitis virus (IBV) -1FS signal.
The MuLV polyprotein Pol is expressed solely as a fusion with the Gag polyprotein by stop codon readthrough at a frequency of 5 %. The biological function of readthrough is hypothesised to be the preservation of a defined ratio (20:1) of Gag and Gag-Pol. Previous studies into the importance of maintaining this ratio have been limited by a failure to test a broad range of readthrough efficiencies (RTEs) without disrupting the functions of the encoded viral proteins. In this work, I used three distinct methods to achieve RTEs from 0.2-100 %. In the first approach, the readthrough signal was targeted by site-directed mutagenesis to generate readthrough-inhibitory changes that were synonymous with respect to the underlying amino acid sequence. I noted a close correlation between diminishing RTEs and declining infectivity, as predicted. Although non-synonymous mutations enabled the preparation of viruses with RTEs up to three-fold higher than wild- type (WT), these lead to detrimental effects on virus protease activity. To circumvent this, a second approach was employed to stimulate RTE up to two-fold, using aminoglycosides that inhibited global translation termination. However, viral replication was unaffected, indicating that this level of stimulation is tolerated by the virus. In the course of this work, I was also able to show that aminoglycosides specifically upregulate readthrough at programmed readthrough sites, and that this effect is additive with that arising from stimulation through mutagenesis of these sites. In a third approach, I developed a two- plasmid system that enabled us to mimic the effect of large increases in RTE from 20 to 100 %. I was able to show that the effect of high Gag-Pol:Gag ratios inside virus-producing cells was generally well-tolerated, although it was cell-type dependent. Using 293T cells, MuLV particles robustly maintained their infectivities up to almost 80 % RTE (a greater than 10-fold increase over WT), despite the fact that virion Gag-Pol levels continued to increase as RTE increased, and that particle numbers diminished substantially with increasing RTE.
In a separate study, I investigated strategies for the purification of ribosomes stalled at the frameshift-promoting RNA pseudoknot of IBV. It is thought that this structure interacts directly with the ribosome, and previous work in the laboratory used cryoEM to study such complexes. Attempts to advance the protocols for this work are described, including the use of nascent-peptide-based purification strategies. Ultimately, it proved impossible to produce sufficient quantities of stalled ribosomes by these methods. The likely explanation for this lies in the obstruction of nascent chain binding to the affinity column by the ribosome or associated factors.
A PDF version of the full PhD dissertation is available at the top of this page. A hard copy is available in Cambridge University library, the record of which lives here.